Catherine Houstis, Christos Nikolaou, Spyros Lalis
University of Crete and Foundation for Research and Technology - Hellas
Sarantos Kapidakis, Vassilis Christophides
Foundation for Research and Technology - Hellas
E-mail: {houstis, nikolau, lalis, sarantos, christop}@ics.forth.gr
* This work is funded in part by the THETIS project, for developing a WWW-based system for Coastal Zone Management of the Mediterranean Sea, EU Research on Telematics Programme, Project Nr. F0069.
The improvement of communication technologies and the emergence of the Internet and the WWW have made feasible the worldwide electronic availability of independent scientific repositories. Still, merging data from separate sources proves to be a difficult task. This is due to the fact that the information required for interpreting, and thus combining, scientific data is often imprecise, documented in some form of free text or hidden in the various processing programs and analytical tools. Moreover, legacy sources and programs understand their own syntax, semantics and set of operations, and have different runtime requirements. These problems are multiplied when considering large environmental information systems, touching upon several knowledge domains that are handled by autonomous subsystems with radically different semantic data models. In addition, there is a widely heterogeneous user community, ranging from authorities and scientists with varying technical and environmental expertise as well as the public.
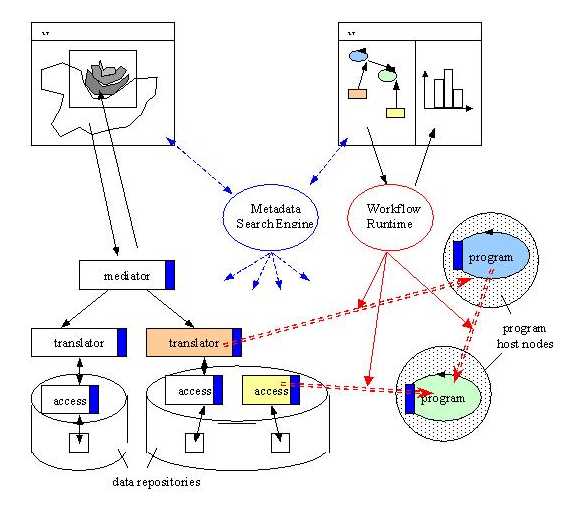
To address these problems we envision an open environment with a middleware architecture that combines digital library technology, information integration mechanisms and workflow-based systems to address scalability both at the data repositories level and the user level. A hybrid approach is adopted where the middleware infrastructure contains a dynamic scientific collaborative work environment and mediation to provide both the scientists and simple users the means to access globally distributed heterogeneous scientific information. The proposed architecture consists of three main object types: data collections, mediators, and programs.
Data collections represent data that are physically available, housed in the systemís repositories. Scientific data collections typically consist of ordered sets of values, representing measurements or parts of pictures, and unstructured or loosely structured texts containing various kinds of information, mostly used as descriptions directed to the user. Each data collection comes with a set of operations, which provide the means for accessing its contents; the access operations essentially combine the model according to which items are organized within the collection and the mechanisms used to retrieve data.
Mediators are intermediary software components that stand between applications and data collections. They encapsulate one or more data collections, possibly residing in different repositories, and provide applications with higher-level abstract data models and querying facilities. Mediators are introduced to encode complex tasks of consolidation, aggregation, analysis, and interpretation, involving several data collections. Thus mediators incorporate considerable expertise regarding not only the kind of data that is to be combined to obtain value-added information, but also the exact transformation procedure that has to be followed to achieve semantic correctness. In the simplest case, a mediator degenerates into a translator that merely converts queries and results between two different data models. Notably, mediators themselves may be viewed as abstract data collections, so that novel data abstractions can be built incrementally, by implementing new mediators on top of other mediators and data collections. This hierarchical approach is well suited for large systems, because new functionality can be introduced without modifying or affecting existing components and applications.
Programs are procedures, described in a computer language that implement pure data processing functions. Rather than offering a query model through which data can be accessed in an organized way, programs generate data in primitive forms (e.g. streams or files). They also perform computationally intensive calculations, so that appropriate recovery and caching techniques must be employed to avoid repetition of costly computations. Last but not least, program components can be very difficult to transport and execute on platforms other than the one where they are already installed, so that they must be invoked remotely.
Every component has metadata associated with it. Thus, registered objects can be located via corresponding meta-searches about their properties. Data collections and mediators bear metadata describing their conceptual schemata and querying capabilities. Programs are described via metadata about the scientific models implemented, the run time requirements of the executable, and the type of the input and output. Metadata are the key issue in an cross-disciplinary system with numerous underlying components: only when we know all significant details about data and programs we will be able to combine them and use them with minimal extra human effort.

This hybrid approach greatly enhances flexibility. On the one hand, mediation is used to offer robust data abstraction with advanced querying capabilities, and to promote incremental development, which is required to build heavy-weight applications such as decision making tools. On the other hand, the collaborative workflow environment is addressing the needs of the scientific community for flexible experimentation with the available system components, at run time. In fact, a workflow can be used to test processing scenaria for a particular user community, in a "plug-and-play" fashion, before embodying them into the system as mediators and applications.